TL;DR
Data cleaning allows us to identify and correctly handle potentially problematic records in our dataset. This post will take you through how to identify a range of potential issues and numerous methods that can be used to deal with such records.
Introduction
Data cleaning is a task that plays a major role in many data analysis projects. Data cleaning is the process of detecting, and then potentially correcting, corrupt or inaccurate records from your data. These records can occur for a range of reasons including data input errors, data missing due to not being recorded or faults in measurement methodologies of technologies (analytic machine errors for example). This is a vast topic with many nuances, in this post we will look to provide an introduction to some of the most common data cleaning tasks you may need to perform.
An initial look
Before we start diving into our data, it is worth spending some time checking our data follows a few basic features. In this post, we will use data from the Mental Health in Tech Survey from 2016.
mht_data <- read.csv("/Users/jonahthomas/R_projects/health_stack/static/files/mental-heath-in-tech-2016_20161114.csv")
#remember, if you have created a project then move the data file into the project directoryThis is a big dataframe and we will not use all of it in this post, instead we will use several variables to examine several different data cleaning techniques. First however, lets look at the dataset as a whole.
Renaming columns
There are a number of things we may want to check including the variable names within our dataframe.
#get the column names then select the first 5
colnames(mht_data)[1:5] ## [1] "Are.you.self.employed."
## [2] "How.many.employees.does.your.company.or.organization.have."
## [3] "Is.your.employer.primarily.a.tech.company.organization."
## [4] "Is.your.primary.role.within.your.company.related.to.tech.IT."
## [5] "Does.your.employer.provide.mental.health.benefits.as.part.of.healthcare.coverage."Using the colnames functions we can examine the column names within our dataframe. The code above shows us the first five column names and we can see they are rather long and cumbersome. We can rename columns using either base R of dplyr functions.
#base r
names(mht_data)[names(mht_data) == "Are.you.self.employed."] <- "self_employed"
#get the names of the columns, filter to the column name you want and then assign it a new value
#dplyr
library(dplyr)
mht_data <- mht_data %>%
rename(employee_n = How.many.employees.does.your.company.or.organization.have.)
#provide the new variable name, followed by the previous variable name
#recheck the data using the below code
mht_data[1:5, 1:5]## self_employed employee_n
## 1 0 26-100
## 2 0 6-25
## 3 0 6-25
## 4 1
## 5 0 6-25
## Is.your.employer.primarily.a.tech.company.organization.
## 1 1
## 2 1
## 3 1
## 4 NA
## 5 0
## Is.your.primary.role.within.your.company.related.to.tech.IT.
## 1 NA
## 2 NA
## 3 NA
## 4 NA
## 5 1
## Does.your.employer.provide.mental.health.benefits.as.part.of.healthcare.coverage.
## 1 Not eligible for coverage / N/A
## 2 No
## 3 No
## 4
## 5 YesVariable type
We may also want to check what type our variables are currently stored in. We can do this using the type_of function.
#check the type of a column
typeof(mht_data$employee_n) ## [1] "character"We can see that the number of employees is currently classified as a character vector, however if we examine the content of this variable we will see there are actually six categories and therefore should be considered a factor variable.
mht_data$employee_n <- as.factor(mht_data$employee_n)
#convert a column to type factor
#examine the levels of a factor
levels(mht_data$employee_n) ## [1] "" "1-5" "100-500" "26-100"
## [5] "500-1000" "6-25" "More than 1000"Above we have used the as.factor function to convert the variable from its current type to the type factor. I have then used the levels function to examine how many levels the variable has and the names of these. We now have our factors however you can see they are not in the correct order (6-25 is in the wrong place). We can change this with the code below.
mht_data$employee_n <- factor(mht_data$employee_n, levels = c("1-5", "6-25", "26-100", "100-500", "500-1000", "More than 1000", ""))
#make the column a factor and set the levels in our chosen order
#examine the levels of a factor
levels(mht_data$employee_n) ## [1] "1-5" "6-25" "26-100" "100-500"
## [5] "500-1000" "More than 1000" ""We can now see our oder is much improved. This is particularly helpful as it means if we graph this factor later the correct order should be followed. Whilst with this column I convert data to a factor, there is also the as.numeric, as.character and as.Date function which you can use as necessary.
Checking for input errors/variability
Often in questionnaire data we may use an open entry field where participants can enter any response they wish. This was the case for the gender column in our dataframe. To see the variety of responses we can use the unique function to see all the unique responses.
#check the number of unique gender responses
length(unique(mht_data$What.is.your.gender.)) ## [1] 72#look at the first 10 unique responses
unique(mht_data$What.is.your.gender.)[1:10] ## [1] "Male" "male" "Male "
## [4] "Female" "M" "female"
## [7] "m" "I identify as female." "female "
## [10] "Bigender"You can see in our 1433 responses there are 72 unique responses. We can then examine the first 10 responses to get a sense for the range of entries provided. Lets say we wanted to categorise these responses into male, female and other we could use the code below.
mht_data <- mht_data %>%
mutate(
gender_category = if_else(substr(tolower(What.is.your.gender.),1,1) == "m", "male", if_else(substr(tolower(What.is.your.gender.),1,1) == "f", "female", "other"))
)
#get the first character of a string, convert it to lower case, check if it is equal to m or f, set the value accordingly
#if neither condition is met set the value to other
#select the gender column
#group by this column
#count the number of entries in each group in the column
mht_data %>%
select(gender_category) %>%
group_by(gender_category) %>%
count() ## # A tibble: 3 x 2
## # Groups: gender_category [3]
## gender_category n
## <chr> <int>
## 1 female 329
## 2 male 1053
## 3 other 51The code above is relatively complex. We start by using the mutate function to create a gender_category variable. We then use the if_else function from dplyr to conditionally create records within the variable. We then use two string manipulation functions, substr and tolower to get the first character from the What.is.your.gender. string and convert it to lower case. If this value equals m then we assign the variable the value male, if it is equal to f we assign it the value of female, and if neither condition is met we assign it the variable other. We then select our newly created column, group_by it and then count it to see the distribution in our sample. We can see we now have three categories.
This method is obviously not perfect (someone could enter a response that starts with an f or m and then provide their gender later in the string) however it is likely the best we can do with the data available.
The above covers a range of techniques that you may want to use when you first load a dataframe into R to check it has the format you require. Next, lets look at how to identify missing values.
Missing values
Missing values can be represented in two main ways in R: either a blank entry in a character string ("" for example) or it has the value NA. In this section we will look primarily at identifying NA values. Lets look at the age column an identify the NA values in the column. (In this dataset, there is actually no NA values in the age column - so I have changed 10% of the values to NA so we actually have something to find!).
set.seed(1234)
library(missMethods)
mht_data<- delete_MCAR(mht_data, 0.1, "What.is.your.age.")
#convert 10% of data to NA completely randomly in the age columnNow we actually have some missing data lets examine it! We can use the is.na function to examine the number of NA values within a column. This generates a vector of true and false values that we can use the sum function on to generate a total number of missing values. We can use a range of iterating functions (such as for loops, or summarise across for example) to apply this to multiple columns.
#use is.na to check if each record is equal to NA and then sum the TRUE values (sum by default only adds TRUE values)
sum(is.na(mht_data$What.is.your.age.)) ## [1] 143We can also use the summary function (we will discuss this more later) and it will count the number of NA’s for us.
#use the summary function on the age column
summary(mht_data$What.is.your.age.) ## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## 3.00 28.00 33.00 34.33 38.75 323.00 143Now we understand the number of NA’s within our data, we need to decide how to deal with them.
Removing NA values
There are three main options when dealing with missing values: removing the entire record with an NA (list wise deletion), remove only records with NA that will be included within your analysis (pair wise deletion - this allows us to keep the maximum amount of data) or imputation of the missing values. The choice is very dependent on the data and field of study.
To remove NA values, we can use the dplyr function na.omit however this removes any row with an NA or missing value (list wise deletion) which in our dataframe removes every row as shown below.
mht_data %>%
na.omit()
#use dplyr na.omit function to drop all rows with NAInstead of this method we can use the drop_na function from tidyr, and when we provide it with a specific column name, we remove all rows with an NA in only the age column.
library(tidyr)
mht_data %>%
drop_na(What.is.your.age.) %>%
head()
#drop rows only with NA in the age columnImputing NA values
There is easily enough information regarding imputing NA values to write an entire blog post (and likely a small book!) but I will try and provide a whistle stop tour. We may chose to impute data rather than remove it if a large amount of our data is missing (removing many rows can significantly reduce our sample size) and this may introduce bias into our analysis. A commonly used method is imputing with a given value (the mean, median or pre-defined value).
The code below imputes the missing data with the mean.
mht_data$What.is.your.age.[is.na(mht_data$What.is.your.age.)] <- mean(mht_data$What.is.your.age., na.rm = T)
#filter so only the columns in the age column that equal NA are selected
#then assign these records the value of the mean of the age columnWe can alternatively use the Hmisc package to impute all our missing data with the median value.
library(Hmisc)
impute(mht_data$What.is.your.age., median)
#replace all NA values in age with the median of the age columnFinally, we could just pick a value for age (40) and replace all our NA values with it.
impute(mht_data$What.is.your.age., 40)
#replace all NA values in age with the value 40All the above methods replace our NA values with different values and can lead to differences in results. Therefore, subject specific knowledge is important to understand the potential impacts of imputing values and the methodology chosen to do this.
Taking the above further, we can also use more advanced predictive techniques (k nearest neighbour classifiers, linear or logistic regression or ANOVA) to use other variables to predict the value of the missing value. As mentioned before, all these approaches have a variety of advantages and drawbacks that need to be balanced based on case specific details.
Is my data missing at random?
You may have been asked to confirm “Is your data missing at random?”. This seems like a relatively complex question however it can be answered quite simply. To answer this we can say is there a difference in missing age data between the gender categories.
We can do this with code similar to the below.
missing <- mht_data %>%
select(What.is.your.age., gender_category) %>%
filter(is.na(What.is.your.age.))
#select age and gender column
#filter to only include where age is equal to NA
count_missing <- missing %>%
group_by(gender_category) %>%
count()
#group by gender and then count them
#convert the count to percentage
count_missing %>%
mutate(percent = n/nrow(missing) * 100)## # A tibble: 3 x 3
## # Groups: gender_category [3]
## gender_category n percent
## <chr> <int> <dbl>
## 1 female 65 23.9
## 2 male 196 72.1
## 3 other 11 4.04not_missing <- mht_data %>%
select(What.is.your.age., gender_category) %>%
filter(!is.na(What.is.your.age.))
#select age and gender column
#filter to only include where age is not equal to NA
count_not_missing <- not_missing %>%
group_by(gender_category) %>%
count()
#group by gender and then count them
#convert the value to percentage
count_not_missing %>%
mutate(percent = n/nrow(not_missing) * 100) ## # A tibble: 3 x 3
## # Groups: gender_category [3]
## gender_category n percent
## <chr> <int> <dbl>
## 1 female 264 22.7
## 2 male 857 73.8
## 3 other 40 3.45We can see from these two tables that the difference in percentage between male, female and other are broadly similar. We could run a statistical test (a t-test) to see if the differences are statistically significant. If they are not significant, you can suggest the data is likely missing at random (or at least, not missing in greater or lesser proportion based on another variable).
Outliers
Outliers are data points that fall outside of the expected range of responses. This can be for a number of reasons including input error (an age of 181 rather than 81), it may be a very high or low value (a patient with a blood pressure of 220/160) or it may be caused by a measurement error (a heart rate monitor malfunctions and records 999 for a few seconds). We will look how to identify such outliers and then how we can deal with them.
Identifying outliers
As mentioned before, we will now return to the summary function. This function gives us the minimum and maximum value as well as the mean and median. Lets examine the age column from our data.
#run the summary function on the age column
summary(mht_data$What.is.your.age.) ## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## 3.00 28.00 33.00 34.32 38.00 323.00 272So we can see from this summary that we have a minimum value of 3 (biologically plausible but realistically not possible) and a maximum value of 323 (not at all possible!). Therefore, we know we have outliers at both ends of our data. We can now check this visually using a histogram.
#use the base r hist function to plot a histogram
hist(mht_data$What.is.your.age.) 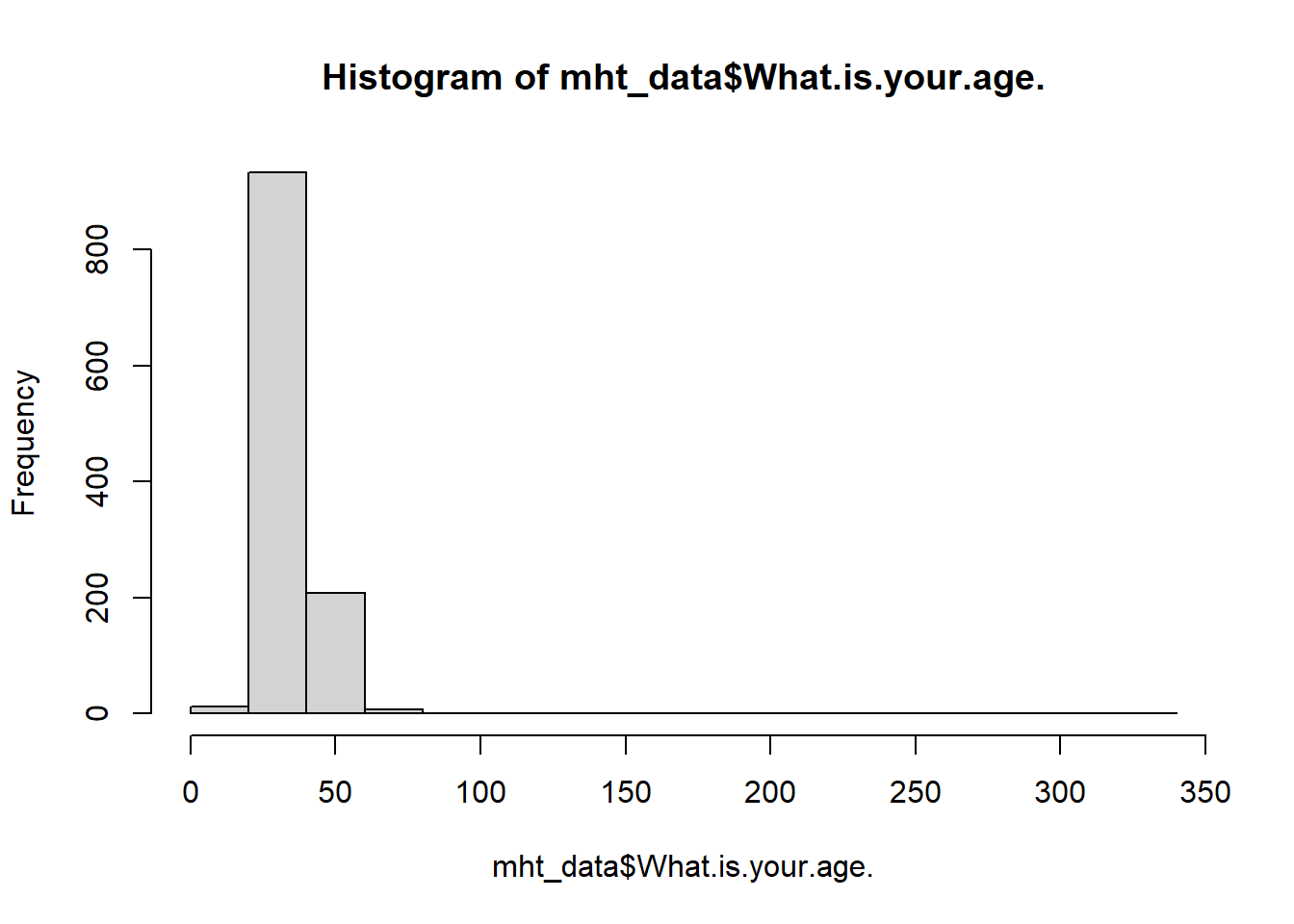
The odd shae of this graph shows our outliers, it appears we may have a couple of low values and one very high value. We can further examine this using a boxplot.
#use the base r boxplot function to plot a boxplot of the age column
boxplot(mht_data$What.is.your.age.) 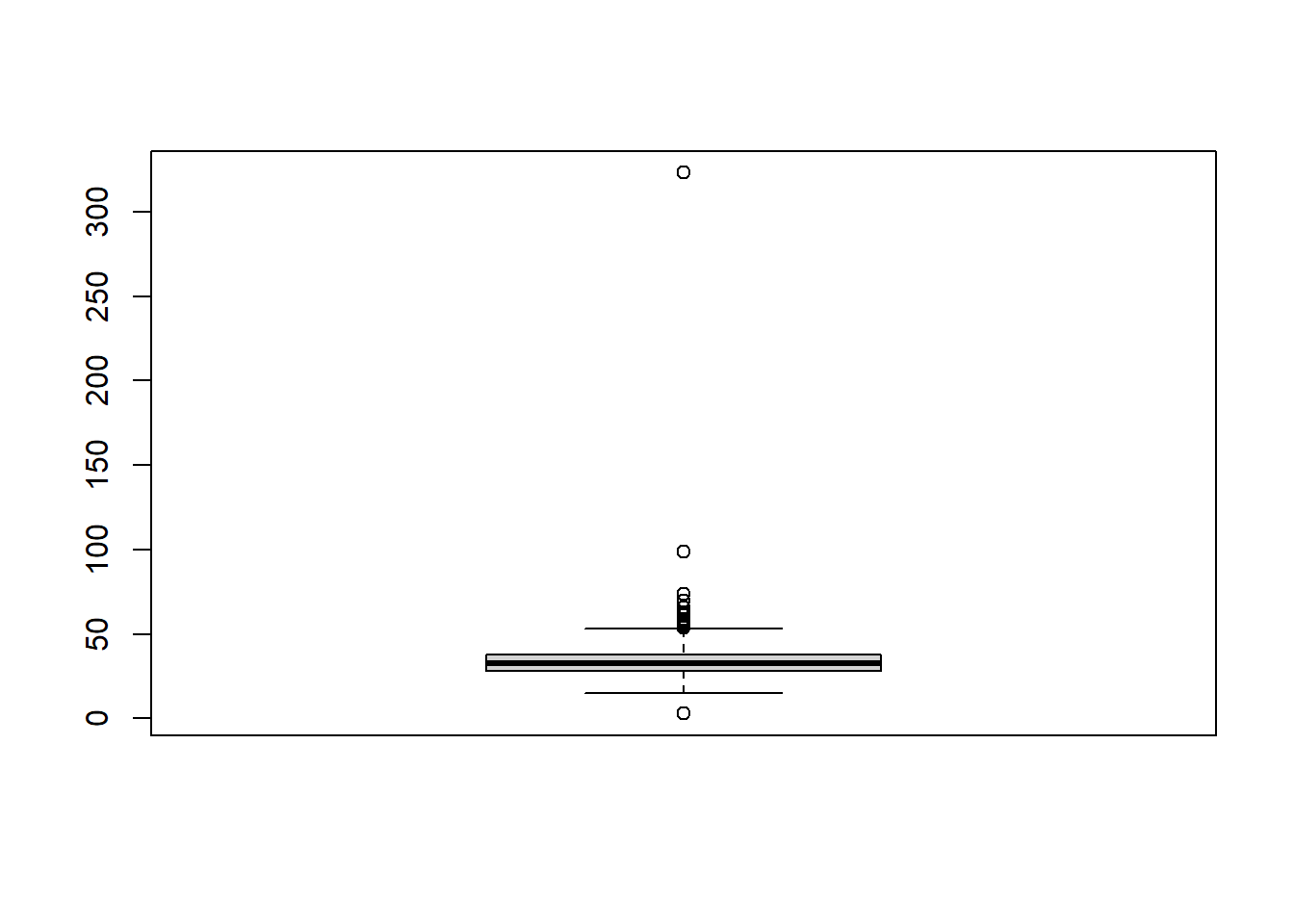
The benefit of the boxplot if we can see the outliers (defined by the hollow circles) and we can identify we have 1 value greater than 300, 1 greater than 100 and one very small value. Personally, this is my preferred method to identify outliers.
Dealing with outliers
Now we understand where our outliers are siting we can look to handle them. One option, is to try and correct these values. In our case, this is not possible but if, for example, paper copies of the records were available these could be double checked and new values entered. However in our case, we are going to remove the values that are not plausible.
#remove values that are not plausible
#count how many rows are left
mht_data %>%
filter(What.is.your.age. <100 & What.is.your.age. >= 18) %>%
nrow()## [1] 1157We can see that by filtering our data to be less than 100 and older than 18 we lose 276 records. We may decide, based on an average retirement age of 70 we could further filter the data to include only records with this value.
#remove values that are not plausible
#count how many rows are left
mht_data %>%
filter(What.is.your.age. <= 70 & What.is.your.age. >= 18) %>%
nrow()## [1] 1155We can see that this only removes an additional 2 records making this choice likely best for this particular dataset.
It is always difficult to decide whether a value is low/high, or an outlier. In our case, we can justify our decision based on biology (people do not work past 100 very often if at all) and then retirement age (it is unlikely anyone works past 70). However, when such justification is not possible a rule of thumb that is often used is ±3 standard deviations. This value is not set in stone and other values can, and have, been used previously.
Other data formats
It is worth noting, whilst the above techniques can be used on a wide range of datasets, other methods are necessary for more specific data types such as time series data (anomaly detection within this field is a vast topic) or log-based data (server logs, app logs etc) require another range of skills not covered in this post. # Conclusions
Data cleaning is a vast area of data science and often a significant part of a data science project (some suggest up to 80-90%). I would argue this is slightly lower for most projects but not all and therefore having a wide range of techniques at your disposable is valuable.
Complete code
mht_data <- read.csv("/Users/jonahthomas/R_projects/health_stack/static/files/mental-heath-in-tech-2016_20161114.csv")
#remember, if you have created a project then move the data file into the project directory
colnames(mht_data)[1:5]
#get the column names then select the first 5
#base r
names(mht_data)[names(mht_data) == "Are.you.self.employed."] <- "self_employed"
#get the names of the columns, filter to the column name you want and then assign it a new value
#dplyr
library(dplyr)
mht_data <- mht_data %>%
rename(employee_n = How.many.employees.does.your.company.or.organization.have.)
#provide the new variable name, followed by the previous variable name
mht_data[1:5, 1:5]
#recheck the data using the above code
typeof(mht_data$employee_n)
#check the type of a column
mht_data$employee_n <- as.factor(mht_data$employee_n)
#convert a column to type factor
levels(mht_data$employee_n)
#examine the levels of a factor
mht_data$employee_n <- factor(mht_data$employee_n, levels = c("1-5", "6-25", "26-100", "100-500", "500-1000", "More than 1000", ""))
#make the column a factor and set the levels in our chosen order
levels(mht_data$employee_n)
#examine the levels of a factor
length(unique(mht_data$What.is.your.gender.))
#check the number of unique gender responses
unique(mht_data$What.is.your.gender.)[1:10]
#look at the first 10 unique responses
mht_data <- mht_data %>%
mutate(
gender_category = if_else(substr(tolower(What.is.your.gender.),1,1) == "m", "male", if_else(substr(tolower(What.is.your.gender.),1,1) == "f", "female", "other"))
)
#get the first character of a string, convert it to lower case, check if it is equal to m or f, set the value accordingly
#if neither condition is met set the value to other
mht_data %>%
select(gender_category) %>%
group_by(gender_category) %>%
count()
#select the gender column
#group by this column
#count the number of entries in each group in the column
set.seed(1234)
library(missMethods)
mht_data<- delete_MCAR(mht_data, 0.1, "What.is.your.age.")
#convert 10% of data to NA completely randomly in the age column
sum(is.na(mht_data$What.is.your.age.))
#use is.na to check if each record is equal to NA and then sum the TRUE values (sum by default only adds TRUE values)
summary(mht_data$What.is.your.age.)
#use the summary function on the age column
mht_data %>%
na.omit()
#use dplyr na.omit function to drop all rows with NA
library(tidyr)
mht_data %>%
drop_na(What.is.your.age.) %>%
head()
#drop rows only with NA in the age column
mht_data$What.is.your.age.[is.na(mht_data$What.is.your.age.)] <- mean(mht_data$What.is.your.age., na.rm = T)
#filter so only the columns in the age column that equal NA are selected
#then assign these records the value of the mean of the age column
library(Hmisc)
impute(mht_data$What.is.your.age., median)
#replace all NA values in age with the median of the age column
impute(mht_data$What.is.your.age., 40)
#replace all NA values in age with the value 40
mht_data<- delete_MCAR(mht_data, 0.1, "What.is.your.age.")
#sets up the data for the exercise
missing <- mht_data %>%
select(What.is.your.age., gender_category) %>%
filter(is.na(What.is.your.age.))
#select age and gender column
#filter to only include where age is equal to NA
count_missing <- missing %>%
group_by(gender_category) %>%
count()
#group by gender and then count them
count_missing %>%
mutate(percent = n/nrow(missing) * 100)
#convert the count to percentage
not_missing <- mht_data %>%
select(What.is.your.age., gender_category) %>%
filter(!is.na(What.is.your.age.))
#select age and gender column
#filter to only include where age is not equal to NA
count_not_missing <- not_missing %>%
group_by(gender_category) %>%
count()
#group by gender and then count them
count_not_missing %>%
mutate(percent = n/nrow(not_missing) * 100)
#convert the value to percentage
summary(mht_data$What.is.your.age.)
#run the summary function on the age column
hist(mht_data$What.is.your.age.)
#use the base r hist function to plot a histogram
boxplot(mht_data$What.is.your.age.)
#use the base r boxplot function to plot a boxplot of the age column
mht_data %>%
filter(What.is.your.age. <100 & What.is.your.age. >= 18) %>%
nrow()
#remove values that are not plausible
#count how many rows are left
mht_data %>%
filter(What.is.your.age. <= 70 & What.is.your.age. >= 18) %>%
nrow()
#remove values that are not plausible
#count how many rows are left